Research
The use and misuse of antibiotics has let to high rates of resistance to these drugs among pathogenic bacteria. Antibiotic resistance can arise through many different mechanisms: including mutations in the drug target, enzymatic inactivation the drug itself and changes to the cell permeability which prevent antibiotic uptake or pump antibiotics out of the cell. However, bacteria can also survive antibiotic treatment without being resistant, but instead entering a dormant, antibiotic-tolerant state during treatment, and reviving and regrow once treatment ends. This phenomenon, termed antibiotic tolerance, can evolve in response to antibiotics and is a potentially major cause of treatment failure and infection recurrence.
To understand how these two important phenomena impact our ability to treat bacterial infections we need to bridge the gap in our understanding of the molecular mechanisms involved and the effect they have on the broader context of the infection. In the lab we take an interdisciplinary approach to studying both antibiotic resistance and antibiotic tolerance, combining molecular microbiology and advanced microscopy methods with evolution studies, pathogen genomics, and analysis of clinical records. We focus in particular on urinary tract infections (UTIs) and their main causative pathogen Escherichia coli , which are the most common reason for prescribing antibiotics in adults. Our ultimate goal is to use a better mechanistic understanding to develop better ways to treat bacterial infections and new strategies to reduce the spread of antibiotic resistance.
Reducing the emergence of antibiotic resistance
Antibiotics are a double edged sword: while they help clear an ongoing infection they also select for resistant pathogens within the patients microbiota, making future infections harder to treat. We are interested in understanding how antibiotic resistance emerges during treatment and how this can be prevented. Using genomic sequencing techniques and machine learning analysis of clinical records from UTI and wound infection patients, we developed an antibiotic prescribing algorithm which cuts the risk of emergence of antibiotic resistance by half.
Bacteria can evolve by randomly acquiring mutations that makes them resistant, but the randomness of the process makes it hard to predict and to avoid. However, sequencing bacterial isolates from patients before and after antibiotic treatments showed that in most patient’s infections resistance was not acquired by random mutations. Instead, resistance emerged due to reinfection by existing resistant bacteria from the patient’s own microbiome. We found that the antibiotic susceptibility of the patient’s past infections could be used to predict their risk of returning with a resistant infection following antibiotic treatment and recommend the best antibiotic for each patient to minimize this risk.
For more information see Stracy et al, Science 2022 and de Nies et al, Nature Reviews Microbiology 2023. For more details about personalised medicine for combatting antibiotic resistance, take a look at this recent post for the British Society for Antimicrobial Chemotherapy website.
Overcoming antibiotic tolerance
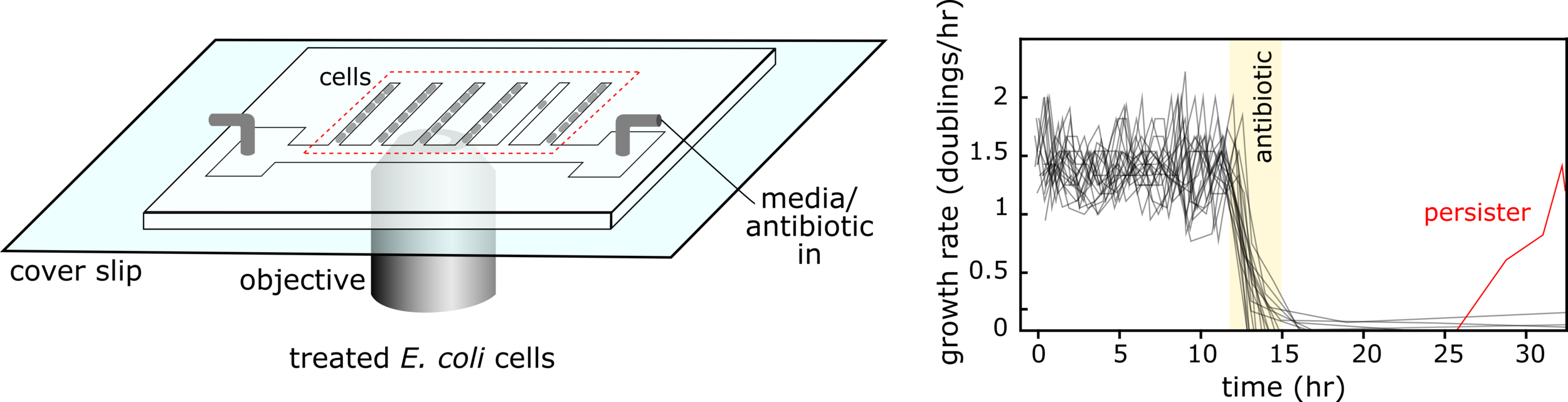
Antibiotics can often fail to clear infections not because the causative bacteria are resistant, but rather because they transiently enter a slow growing ‘persister’ state in which they can survive antibiotics and revive after treatment ends leading to infection recurrence. Furthermore, tolerance-conferring mutations can rapidly evolve in response to treatment. We are working on new way to eradicate these ‘sleeping’ bacteria to help stop infections recurring and reduce the number of antibiotic courses that patients need to take.
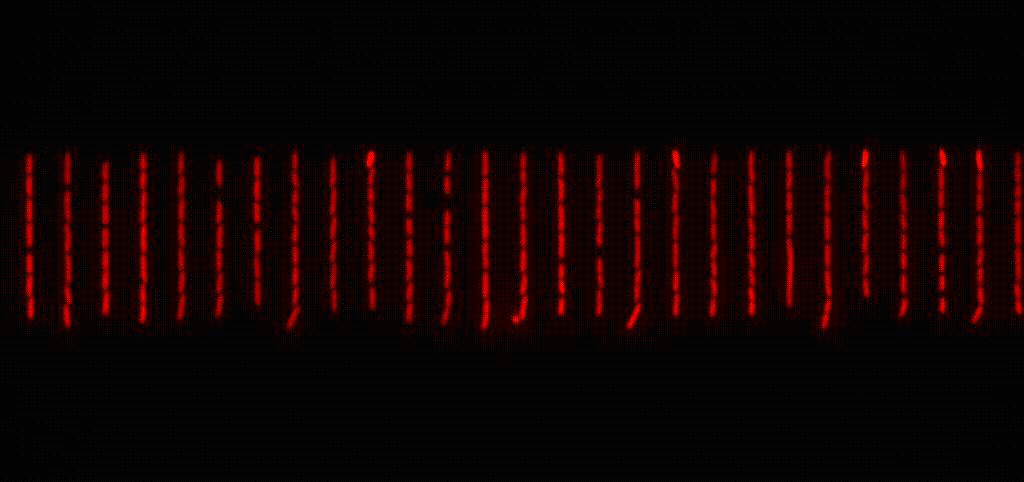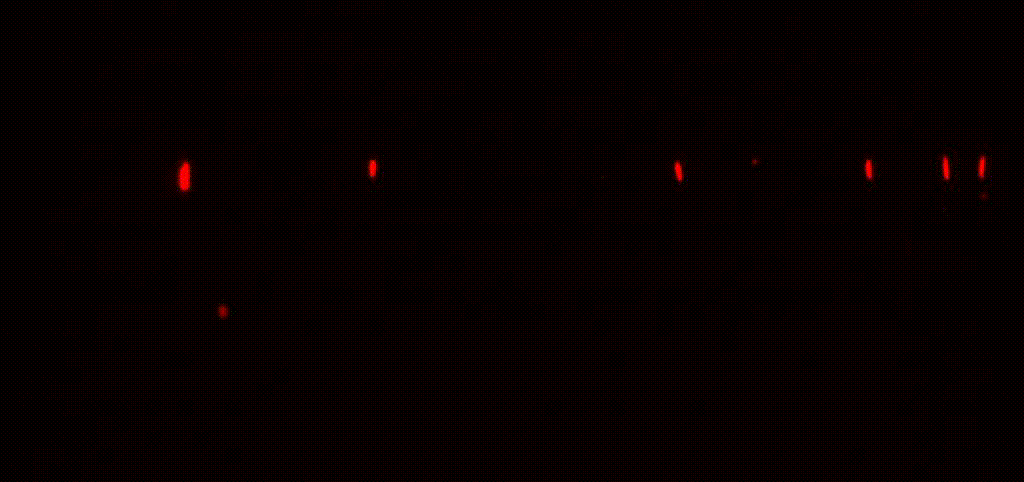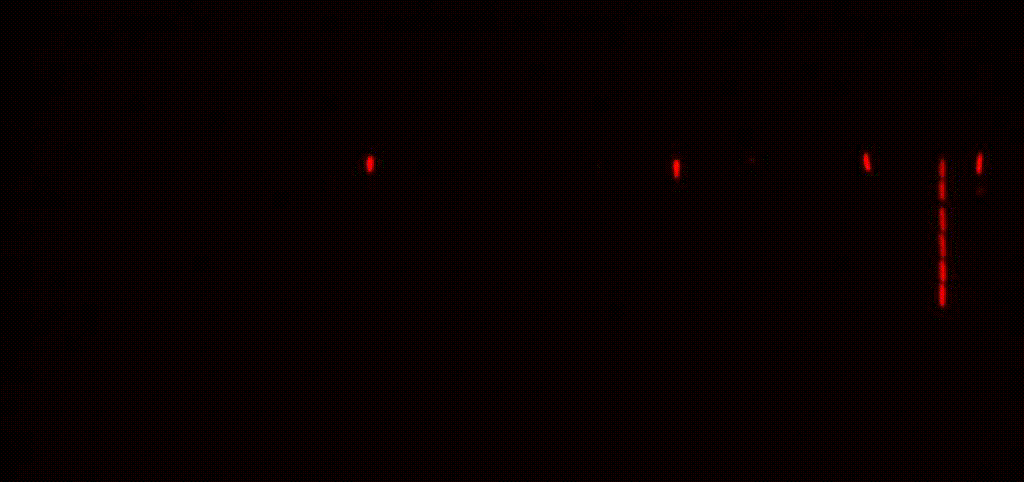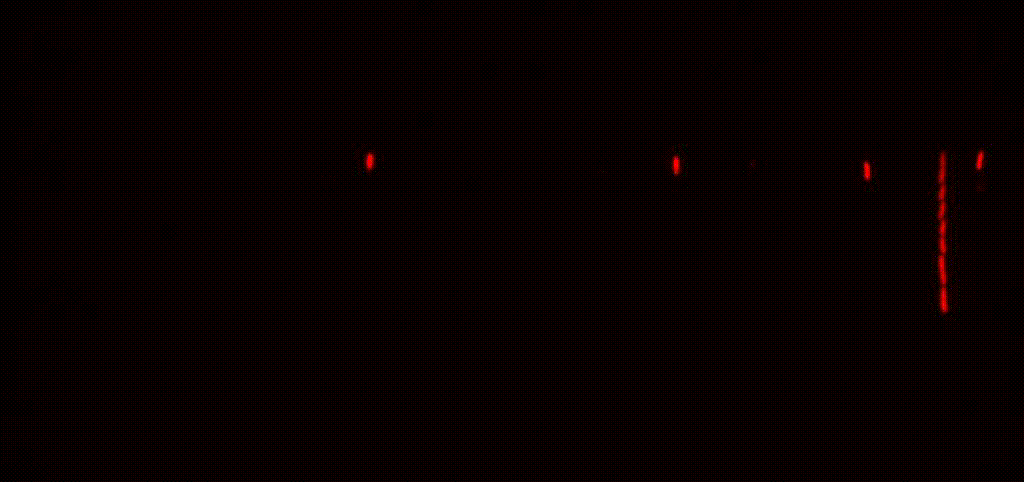
We study antibiotic tolerance using a variety of techniques including laboratory evolution and advanced microscopy methods. We image bacteria in microfluidic devices before, during and after antibiotic treatment to identify surviving persister cells. This allows us to explore the physiology of dormant antibiotic-tolerant cells to help unravel why they survive treatment.
How do antibiotics kill bacteria?
We are also interested in the fundamental molecular biology of how antibiotics work and how they corrupt their target proteins to kill cells. In particular we are interested in how the efficacy of antibiotics depend on the bacteria’s physiological state and its environmental conditions. One of the ways we study this is by directly imaging and quantifying antibiotic-target binding inside cells using single-molecule fluorescence microscopy combined with single particle tracking.
For more information see Stracy et al, Nucleic Acids Research 2019, Stracy et al, Nature Communications 2016.